
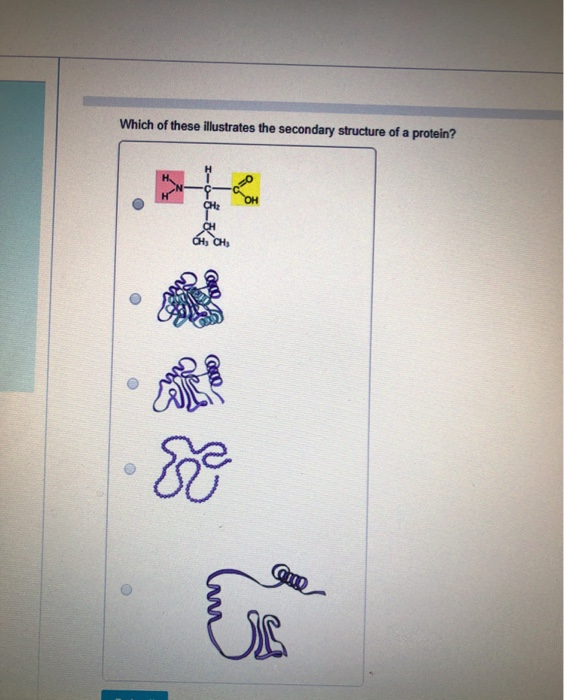
Hydrogen Bond Energy dihedral angle probabilities Knowledge-based protein secondary structure assignment. But what is the problem with doing this?Ģ4 Relative solvent accessibility (Range = 0-1)Ģ6 STRIDE: secondary STRuctural IDEntification Biopolymers Dec 22(12):Ģ3 An aside: ACC from DSSP ACC = water exposed surface (Å2). Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Used to define chirality (structure code '+' or '-'). **ALPHA: virtual torsion angle (dihedral angle) defined by the four Cα atoms of residues I-1, I, I+1, I+2.

Used to define bend (structure code 'S'). (Not used for structure definition) **KAPPA: virtual bond angle (bend angle) defined by the three Cα atoms of residues I- 2, I, I+2. Bifurcated HBsĢ0 DSSP Sample Output **TCO: cosine of angle between C=O of residue I and C=O of residue I-1. There are two columns for each type of H-bond, to allow for bifurcated H-bonds. 3,-1.4 means: if this residue is residue i then N-H of i is h-bonded to C=O of i-3 with an electrostatic H-bond energy of -1.4 kcal/mol. A simple way: angle-distance hydrogen bond assignment: A hydrogen bond is assigned when: 1. Backbone geometry (main-chain dihedral angles) Hydrogen bond patterns “Knowledge-based protein secondary structure assignment”. What are the two main structural properties when we talk about secondary structures? 2. first strandĐ Parallel 1 anti-parallel -1 They are assigned by crystallographers, but how? Will come back to this later…… Types of helices and sheets: Helix: Right-handed alpha (default) 1 Right-handed omega 2 Right-handed pi 3 Right-handed gamma 4 Right-handed 310 5 ……………… Sheets: Sense of strand with respect to previous strand in the sheet. It has functional implications Useful in protein structure comparison and protein structure prediction - Some protein structure alignment programs use SSE (secondary structure element) - In protein threading, secondary structures are used to define “cores”, more later… It is an intuitive means of visualizing and understanding protein structures lysozyme CAF Andersen, and B Rost (2002) “Secondary structure assignment”ĥ Secondary Structure Annotations from PDB File DSSP and STRIDE Topic 9 Chapter 19, Du and Bourne “Structural Bioinformatics”Ī key step in protein classification -class, fold….


 0 kommentar(er)
0 kommentar(er)
